Introduction
The OpenAPI project aims to help people connect with
data. OpenAPI is a software architecture for wrapping component
pieces of data analysis—datasets, programming scripts,
etc.—in reusable modules. These simple modules can be
combined in pipelines which describe more complex data analysis
workflows. Modules and pipelines are created in XML, and
executed in software called a glue system. This report details
the improvements made to the OpenAPI architecture between
versions 0.1 and 0.3.
Version 0.1 of the OpenAPI architecture introduced the
OpenAPI module XML specification for wrapping data analysis
scripts. The XML specification for modules was not well-suited
to wrapping scripts which had not been written with OpenAPI in
mind. This report describes the changes made to the OpenAPI
architecture in version 0.3. These changes include the
introduction of 'vessel' elements for describing inputs and
outputs to OpenAPI modules which improve the authoring of module
and pipeline XML. This report also describes the introduction of
a module 'host' machine which allows OpenAPI to address issues
concering software dependencies; these issues affect the
portability and reproducibility of data analysis
workflows. Version 0.3 of the OpenAPI architecture also
introduces XML Schema for module and pipeline XML, allowing
module and pipeline authors to validate their files.
This report demonstrates how the changes introduced in
OpenAPI v0.3 make it much easier for module authors to wrap
'wild' data analysis scripts in OpenAPI modules. This report
provides examples of how OpenAPI can be used to execute data
analysis code from external scripts, and even from blog
posts. The report demonstrates how executing 'wild' source
scripts using OpenAPI maintains a connection to the original
script sources, while also allowing a module or pipeline author
to extend or adapt the scripts she wraps. Wrapping data analysis
scripts in module XML also allows a module author to archive
versions of a script against the sometimes ephemeral nature of
online resources.
The OpenAPI pipelines and modules described in this report
were all created, tested and executed in conduit, a prototype
OpenAPI glue system implemented as an R package. The conduit
package source code is available online at
https://github.com/anhinton/conduit. The conduit
package was also improved as part of this project: these changes
were implemented in version 0.3 of conduit, which available
for download at
https://github.com/anhinton/conduit/releases/tag/v0.3.
This report may give the impression that OpenAPI and
conduit have made the leap from version 0.1 to 0.3 without any
mention of version 0.2. Many of the changes in this report were
implemented in version 0.2 of OpenAPI and conduit (Hinton, 2015b). However, as
version 0.3 represents a more fully realised implementation of
the changes described here, only version 0.3 is mentioned
within.
Outline
The following outline should serve as a map to the various
sections of this report:
-
Background: The first section of
this report provides some context for the aims of the OpenAPI
project. This section also describes the composition of the
OpenAPI architecture, and how its pieces interact.
-
Problems in
OpenAPI version 0.1: the second section of this report
describes the problems identified in the XML specification of
OpenAPI v0.1. This section also describes the difficulties of
wrapping data analysis scripts which were not written
specifically for OpenAPI, i.e. almost every data analysis
script in the 'wild'. The XML examples in this section were
written using OpenAPI v0.1.
-
Changes in
OpenAPI version 0.3: this section details the changes in
the OpenAPI XML specification found in version 0.3. This
section demonstrates improvements over OpenAPI v0.1 using XML
examples. Each change to OpenAPI is followed by a module or
pipeline example to illustrate the improvement.
-
Running 'wild'
source scripts: This section provides examples of how the
improvements in OpenAPI v0.3 make it easier to wrap source
scripts, including those found embedded in other
documents. These examples also demonstrate how OpenAPI v0.3
makes it possible to subset and modify 'wild' data analysis
scripts while preserving connections to original scripts. The
examples in this section demonstrate the changes and
improvements made in OpenAPI v0.3.
-
Technical
requirements: Details of the system used to execute the
pipeline and module examples contained in this report.
-
Pipeline
examples: Full pipeline and module examples used in the
report, including code for executing the pipelines in
conduit v0.3, and final output.
Background
As the world collects and makes available more and more data
it is increasingly important for people to be able to access,
understand and use this data to make decisions in their
lives. Government initiatives like the New Zealand Government
Open Access and Licensing framework (NZGOAL), and data.gov.uk
have made more government and public data available online
(Department of Internal Affairs,
2011; Cabinet
Office, 2010).
Almost all newly available data is provided online digitally,
in a variety of proprietary and open file formats; these files
can be read into various commercial and open software
applications where the user can manipulate the data, perform
analyses on them, and produce various summaries and graphics
describing the data. A key challenge in helping people to
connect with data is that even with freely accessible data and
freely available software tools the data user still needs a host
of skills to make use of the data, and of the tools. Hinton and
Murrell (2015a) suggest
that, as well as access to data, the following are required for
people to fully connect with data:
- domain knowledge: a data user cannot make use of data if she
does not have a contextual understanding of where the data comes
from and what it means;
- data science skills: a data user must know what analytical
tools are available and how they can be used to examine data
sets;
- statistical graphics skills: a data user must know what
graphical tools are available, and how they can be used to
explore the characteristics of the data;
- graphical design skills: a data user must know how to
produce meaningful graphics which do not mislead the reader, and
which meet graphical design and presentation norms.
It is understandably very rare to find someone who is the
master of all of these skills, however, and it is perhaps too
demanding to expect an everyday person to be expert in even one
of these categories. One solution to this problem is to design
software which provides many of these skills for the user, and
which allows users to share what skills they have with other
users. OpenAPI takes a different approach; as described in
Hinton (2015a) the OpenAPI
architecture is designed to help solve this problem by
being:
- modular,
- reusable,
- shareable,
- extensible,
- open and free,
- not overly technical,
- not monolithic, and,
- compatible with reproducible research.
Perhaps most importantly, the OpenAPI architecture is
intended to make it easy to incorporate existing resources, like
data analysis scripts, without the authors of these scripts
having to be aware of OpenAPI at all.
The OpenAPI architecture
The OpenAPI architecture consists of XML specifications for
'module' and 'pipeline' files: module XML files wrap scripts of
programming code which describe tasks in data analysis; pipeline
XML files describe how to connect and combine the inputs and
outputs from several modules as a data workflow. The OpenAPI
architecture also specifies requirements for software which can
create, interpret, and execute modules and pipelines; an
instance of such software is called a 'glue system' (Hinton & Murrell,
2015a).
A <module> is the smallest piece in the
OpenAPI system. A module XML file wraps a data script, specifies
the platform or language in which this script can be executed,
and names the inputs required and the outputs produced by this
script (Hinton & Murrell,
2015a).
A <pipeline> XML file describes how a
module's outputs are to be connected to the inputs of other
modules; the output of one module can be provided as the input
of another module through a <pipe>
element. Executing a pipeline results in the execution of all
the wrapped module scripts contained within (Hinton & Murrell,
2015a).
Modules and pipelines are executed by a software 'glue
system'. An OpenAPI glue system is responsible for ensuring that
a module's inputs are satisfied, and that its outputs can be
resolved. The glue system is responsible for providing one
module's output as another module's inputs as described by a
pipeline XML file (Hinton
& Murrell, 2015a). The conduit package is a
prototype OpenAPI glue system written in R. Version 0.1 of
conduit (Hinton &
Murrell, 2015b) was released in February 2015.
The following is a simple example of a pipeline, written in
OpenAPI v0.3. First, the module XML file, 'module1.xml':
<?xml version="1.0"?>
<module xmlns="http://www.openapi.org/2014/" language="R">
<description/>
<source>
<script><![CDATA[x <- rnorm(10)]]></script>
</source>
<output name="x">
<internal symbol="x"/>
<format formatType="text">R numeric vector</format>
</output>
</module>
This module wraps a source script which produces a vector of
ten random numbers in R, called 'x', and labels this as a module
output. A second module XML file, 'module2.xml':
<?xml version="1.0"?>
<module xmlns="http://www.openapi.org/2014/" language="R">
<description/>
<input name="numbers">
<internal symbol="numbers"/>
<format formatType="text">R numeric vector</format>
</input>
<source>
<script><![CDATA[plot(numbers)]]></script>
</source>
<output name="Rplots.pdf">
<file ref="Rplots.pdf"/>
<format>PDF file</format>
</output>
</module>
This module requires an input, called 'numbers'. The module
source script creates a plot of this input in R, and labels the
PDF file of the plot created as an output, called 'plot'. The
following pipeline XML connects these two modules:
<?xml version="1.0"?>
<pipeline xmlns="http://www.openapi.org/2014/">
<description/>
<component name="module1" type="module">
<file ref="module1.xml"/>
</component>
<component name="module2" type="module">
<file ref="module2.xml"/>
</component>
<pipe>
<start component="module1" output="x"/>
<end component="module2" input="numbers"/>
</pipe>
</pipeline>
This pipeline loads each module as a component,
and connects the 'x' output from 'module1' to the 'numbers' input
from 'module2' using a pipe element. When this
pipeline is executed by a glue system 'module1' will be executed
first; its output, 'x', will be made available to 'module2', which
will then be executed.
The OpenAPI architecture attempts to improve the
accessibility and reusability of data sources and computer
scripts, particularly scripts for working with data, by
providing multiple methods of contributing to a solution, at
multiple levels of skill and expertise. The next section
describes some of the problems with the version 0.1 of the
OpenAPI architecture.
Problems in OpenAPI version 0.1
Version 0.1 of the OpenAPI XML specification of pipeline and
module XML files was described in Hinton and Murrell (2015a). A key feature of the
OpenAPI architecture is that it should allow users to wrap
scripts—especially those authored by someone
else—without having a high knowledge of the script's
programming language. An ideal situation is that a module author
should only have to know the names and formats of the resources
that a script requires in order to be executed (its inputs), and
the names and formats of the resources it produces which the
author may want to consume or retrieve (its outputs). Most
existing scripts—'wild' scripts—will not have been
written with OpenAPI in mind, and so the OpenAPI architecture
must be able to cope with this.
In this section we will describe several problems with OpenAPI
version 0.1, including:
This specification was implemented in version 0.1 of the
conduit package. This implementation suffers from problems
with:
- Specifying module
<input> and
<output> elements
- Specifying module
<source> elements
- Meeting module dependencies
- Validating module and pipeline XML
We will then describe how these problems make OpenAPI version
0.1 unsuitable for transparently wrapping and executing 'wild'
data scripts by way of two examples.
The module XML specification used in OpenAPI version 0.1
requires that module <input> and
<output> elements have a 'name' and a 'type'
attribute; type can be either "internal" or "external". In
the following code we see an example of an internal-type input
and an external-type output:
<module>
<platform name="R"/>
<input name="a" type="internal"/>
<source><![CDATA[data <- myFun(a)
png("example.png")
myPlot(data)
dev.off()
]]></source>
<output name="dataplot" type="external" ref="example.png"/>
</module>
An internal-type input or output is an object which is
particular to the module's platform—the above example is
executed in the "R" platform. A platform's internal objects should
be able to be referenced by a symbol. An object is internal to a
platform if it can be referenced and accessed by its symbol within
the platform and without having to pass the object through the
filesystem or some other storage or transmission format. In R
these internal objects are common R objects, e.g. vectors and data
frames; in Python these are common objects like strings or
lists.
In the above example the input specification indicates that
the module source script requires there to be an R object
available with symbol "a"; the glue system will also use the
name "a" to reference this input in pipelines.
An external-type input or output is independent of the
module's platform. In OpenAPI version 0.1 these objects are
files on the local filesystem. A module with an external-type
output will produce a resource at the location given by the
'ref' attribute; this location could be an absolute file
location or a relative file location. A relative file location
indicates the file produced will be created relative to the
output location of the module when it is executed, the details
of which are implemented by a glue system (Hinton & Murrell, 2015a);
'path' attributes are not defined for external-type outputs. A
module with an external-type input indicates that the module
source scripts expect there to be a resource available on
execution, the location of which is available to the scripts
using the symbol indicated by the 'name' attribute; this
resource location is provided by the glue system when executing
a pipeline which connects this input to an external-type module
output.
In the above example the output specification says that the
module source will produce a file object at relative file location
"example.png"; the name "dataplot" will be used by a glue system to
reference this output in pipelines.
Names and symbols
The first problem with this specification for input and
output types is the conflation between an internal-type input's
name, as it is referenced by a glue system, and its symbol. A
module author is forced to use the same value for the input's
'name' as the symbol used in the source scripts to access this
resource. This restriction can be seen in the module XML example
above: the module has an input named "a", and the module's
source script refers to a symbol, "a". If the module author
wanted to call her input something more meaningful in the
context of her pipeline, e.g. "cleaned_dates", she would
have to change the symbol reference throughout the source
scripts to "cleaned_dates":
<module>
<platform name="R"/>
<input name="cleaned_dates" type="internal"/>
<source><![CDATA[data <- myFun(cleaned_dates)
...
]]></source>
</module>
Alternatively, she could prepend the source script with a line
assigning her preferred name to the symbol used in the source
script, for example:
<source><![CDATA[a <- cleaned_dates
data <- myFun(a)
...
]]></source>
Now the author is free to name her module input
"cleaned_dates", but she must name it this, or alter the
source script again. This limitation restricts the reusability of
modules, and forces the author to have knowledge of language of
any source scripts she might wish to use.
Module authors are similarly constrained in naming
internal-type module outputs; the 'name' specified in the output
element must match a symbol available after the execution of the
source scripts. This imposes the same restrictions as mentioned
above: a module author must either conform to the symbol names
given in the source script in naming internal-type module outputs,
or she must modify the source scripts so as to ensure a symbol
with her preferred name exists.
Passing external objects into modules
A second issue involves the use of external-type module
inputs. OpenAPI version 0.1 makes external object resources
available to module source scripts by creating an object which
contains the resource location before the execution of the
module's source script. If a module needs a CSV file for script
execution, the script must refer to the CSV file location using
a symbol matching the input's 'name' when it is accessed in
code.
As an example, the following R script demonstrates how a script
author might normally refer to a CSV file found at resource
location "bigdata.csv":
if (file.exists("bigdata.csv")) {
data <- read.csv("bigdata.csv")
}
If a module author intended for the resource location used in a
module script to be provided by another module's external-type
output she must refer to the resource location using a symbol
matching her module's external-type 'name' attribute, for
example:
<module>
<platform name="R"/>
<input name="csvfile" type="external"/>
<source><![CDATA[if (file.exists(csvfile)) {
data <- read.csv(csvfile)
}]]></source>
</module>
On execution of a pipeline including this module the conduit
glue system resolves the external-type input resource by
prepending the module's source scripts with an assignment to the
symbol given by 'name':
csvfile <- "bigdata.csv" # prepended by glue system
## original source script follows ##
if (file.exists(csvfile)) {
data <- read.csv(csvfile)
}
This makes writing modules with external-type inputs an
unnecessary burden on the module author as she must carefully
alter the source script. Even if the author has knowledge of the
language in which the script is written it is likely that the
changes required to alter a script in this fashion will not be
typical of how scripts in this language are composed.
Starting with an external input
A third problem with the OpenAPI v0.1 module specification is
the difficulty of running a module with an external-type input
in isolation; an equivalent problem is running a pipeline where
the first module requires an external-type input. An
external-type <input> element does not
contain any information about where the file resource can be
found; this location is resolved by the glue system when a
pipeline is executed; an external-type module input can only be
provided by another module's external type module output. In a
pipeline the originating module's output is connected by a pipe
to the consuming module's input. The originating external-type
output contains a 'ref' attribute, which the glue system uses to
resolve the location of the external resource. As external-type
module inputs do not contain 'ref' attributes the glue system
has no way of resolving a resource location from module XML
alone.
Consider the following module, which requires a CSV file as an
external-type input, and produces an R data frame as an
internal-type output:
<module>
<platform name="R" />
<input name="csvfile" type="external">
<format>CSV file</format>
</input>
<source>
<![CDATA[bigdata <- read.csv(csvfile)]]>
</source>
<output name="bigdata" type="internal">
<format>R data frame</format>
</output>
</module>
If a module author wanted to supply a local file,
'/home/bigauthor/data/bigdata.csv', as an input to this module she
cannot do so using the module XML available. The author could
provide a module with no inputs or sources which names the file
location as a 'dummy' external-type output:
<module>
<platform name="R" />
<output name="bigdata.csv" type="external" ref="/home/bigauthor/data/bigdata.csv">
<format>CSV file</format>
</output>
</module>
The module author can execute her new module by executing a
pipeline connecting these two modules. However she cannot easily
execute her module in isolation. To execute her module on its
own she could remove the external-type input and explicitly
reference the CSV file in her module source script:
<module>
<platform name="R" />
<source>
<![CDATA[bigdata <- read.csv("/home/bigauthor/data/bigdata.csv")]]>
</source>
<output name="bigdata" type="internal">
<format>R data frame</format>
</output>
</module>
This module can now be run in isolation. However it is now only
usable at the start of a pipeline, and cannot receive other
module outputs as an input. Any subsequent user of the module will
have to edit the module's source script to change data sources,
rather than altering the module inputs. This has the undesirable
effect of solving the problem of starting a pipeline from a data
file by sacrificing the reusability of the module.
Files, URIs and URLs
Though we have demonstrated some of the issues with using
external-type module outputs to refer to files on the local
filesystem, the implementation of the external-type module
outputs in conduit version 0.1 actually treats the value of
'ref' as a uniform resource identifier (URI). The conduit
package assumes this URI will use the file:// scheme, however,
no checking of scheme is done by conduit, and the value of an
output's 'ref' is passed directly to any module for which a pipe
connects this output to an input. As described above, conduit
ensures that a symbol referencing the location given by the
output's 'ref' is created prior to the execution of the source
scripts; the script author is responsible for deciding how to
handle this resource location, and whether to handle it using
file://, http://, https://, some other URI scheme, or even as a
custom character string. A glue system cannot determine what
scheme is intended by an external-type output's 'ref' without
parsing the value given.
Therefore, although Hinton and Murrell (2015a) did not document this
fact, it is possible in conduit version 0.1 to provide URLs as
external-type module outputs, and for these to be consumed by
external-type module inputs. For the same reason mentioned above
it is not possible to execute a module which requires an
external-type input referencing a URL in isolation, nor to begin
a pipeline from a URL data source without a dummy module naming
a URL output.
Specifying module source elements
The module XML specification implemented in OpenAPI version
0.1 uses a 'ref' attribute in its <source>
elements in a similar fashion to how it is used in module output
elements: 'ref' provides a URI for a resource which contains a
source script to be executed. As with module outputs the
resource location provided by 'ref' is assumed to use the
file:// scheme by default, but support is also included for
http:// and https:// schemes. As with external-type outputs, a
glue system cannot determine which scheme is used by a source's
'ref' attribute without parsing the value given.
Further, the specification for source elements is inconsistent
between source elements which reference a script using the 'ref'
attribute and source elements which contain the script inline as
the element's content. It is possible for a source element to have
both a 'ref' attribute, and to contain an inline script, as seen
here:
<source ref="cleanData.R">
<![CDATA[x <- runif(100, 1, 100)]]>
</source>
The source script referenced by "cleanData.R" might assign an
entirely different value to the symbol 'x'; it would then be up to
a glue system to decide how to handle this situation, as it is not
obvious from the specification which script should take
precedence. A module author would therefore be uncertain of the
result of executing a module with such a source.
Meeting module dependencies
The current XML specification for OpenAPI modules does not
include any explicit mechanism for addressing the system
dependencies of the module's source scripts. The implementation
requires the user to set the module's source language, called it's
'platform', using the <platform> element as in
the following example:
<module>
<platform name="R"/>
...
<source><![CDATA[library("rgl")
....
]]></source>
...
</module>
If the glue system used to execute this module supports the
"R" language, the source scripts wrapped by the module will be
executed in an R environment. In the above example the
rgl package (Adler,
Murdoch & others, 2015) is attached at the beginning
of the source script; if the rgl package has not
been installed in the glue system's R environment this module
will likely fail to execute. One possible solution to a missing
package in R (or a missing library in Python, etc.) is to change
the source script to guarantee the package is available as
in:
<module>
<platform name="R"/>
...
<source><![CDATA[if (!require("rgl")) {
install.packages("rgl", lib = Sys.getenv("R_LIBS_USER"),
repo="http://cran.stat.auckland.ac.nz")
}
require("rgl")
....
]]></source>
...
</module>
Now the module source script checks to see if the
rgl package is installed, and installs it if
not. However, the rgl package itself has system
dependencies which, if not met, will prevent the installation of
the rglpackage. The OpenAPI module specification
does not provide a mechanism for checking for system
dependencies in such cases.
It is also very likely that module authors will wrap source
scripts which depend on specific versions of software. OpenAPI
v0.1 does not provide a mechanism for specifying which version
of a language, or its associated libraries and packages, should
be installed. Hinton and Murrell (2015a, 'Discussion') suggest
that the OpenAPI XML specification could include a mechanism for
running a module's source scripts on a specific host machine;
this host could be set up with the module's exact software
requirements, and be made available to module users. Support for
host machines is not included in OpenAPI v0.1 or in conduit
v0.1.
Validating module and pipeline XML
Although the XML specification was described in detail by
Hinton and Murrell (2015a) only an informal test
for the validity of an OpenAPI pipeline or module XML file
exists: if a pipeline or module can be read and executed by the
conduit package it is assumed to be valid. It is desirable to
have a more formal specification and test of validity for module
and pipeline XML files in the form of an XML Document Type
Definition (DTD) and/or XML Schema to ensure the consistency of
OpenAPI documents.
Running 'wild' source scripts
The OpenAPI architecture aims to make it easy to wrap and
execute 'wild' data analysis scripts in module XML. However,
several of the problems described above make this very difficult
in OpenAPI v0.1. We will give two examples which highlight the
difficulties in wrapping 'wild' scripts.
The first example demonstrates how a simple R script found in
the 'wild' might present a challenge to an OpenAPI module
author. This script was published on the University of
Auckland's Statistics 330 website as supplementary code to a
lecture on R graphics (Klaere,
2015). The first 21 lines of the script are:
##############################
### STATS 330/762 - Lecture 3: Graphics
### 23/07/2015
### Dr. Steffen Klaere
### Supplementary Code
### NEEDS: hb1.csv and fever_data.csv
##############################
### Change working directory
# Adapt to your own needs
setwd("<Your Directory>")
### Packages
library(R330)
###################################
### First example, Exchange rates
###################################
exchange <- read.csv("hb1.csv")
exchange[,1] <- as.Date(sapply(exchange[,1],function(x){paste(1,x)}),format="%d %B %Y")
plot(USD~month,data=exchange,type="l",lwd=4,col="steelblue",xlab="Date",ylab="Exchange rate",main="Exchange rate between US$ and NZ$")
As Klaere has helpfully indicates on line six, this script
requires two files: 'hb1.csv' and 'fever_data.csv', both
available from the course website; the first of these files is
called on explicitly by its filename in line 19. These files are
obvious candidates for module inputs. However, as mentioned
above, there is no straightforward way of describing file inputs
to an OpenAPI module—file inputs to modules must be the
outputs of other modules, or the module source script must be
modified to adapt to file locations. The module author is faced
with both editing the source script and creating dummy
module outputs simply to provide files to a script. Of course,
she can simply leave the script as is, and make sure the files
are in the right file location, restricting her pipeline's
reusability and modularity. None of these options provides a
simple way of wrapping a script for inclusion in a pipeline, or
even for executing the script on its own.
If even this simple example of wrapping a source script in an
OpenAPI module seems difficult, then wrapping other common
'wild' data scripts proves even more difficult. For example, it
is common to find data analysis scripts, in R, Python, and other
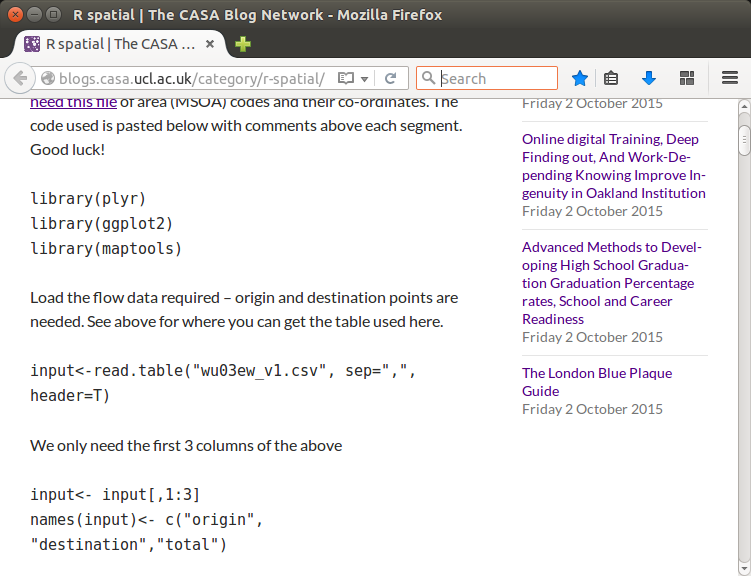
languages, embedded in blog posts, as in Figure 1.
In the blog post shown in Figure 1
(Cheshire, 2015) we see R
code chunks mixed with descriptive text. It would not be
possible to execute the entire blog post as an R script; if an
author wanted to wrap this script in an OpenAPI module she must
select only the parts of the blog post which contain executable
R code. Even then the author must still arrange for the data
file, 'wu03ew_v1.csv', and any other data dependencies, to be
resolved as module inputs with all the same difficulties
mentioned above.
In the next section we will describe how the problems and
limitations in OpenAPI v0.1 have been addressed in OpenAPI
v0.3.
Changes in OpenAPI version 0.3
One of the main aims of this project was to improve the OpenAPI
XML specification to deal with the problems detailed in the
previous section. This project has made improvements to the XML
specification for modules and pipelines by:
- Introducing vessels to reference resources in module
inputs, outputs, and sources, and in pipeline components;
- Refactoring module platform elements into module
'language' attributes;
- Adding host specification to modules;
- Specifying XML Schema for valid pipeline and module
XML.
Vessels
As described in the previous section, the way module inputs,
outputs, and sources were specified was inconsistently designed
in OpenAPI v0.1. Module input and output elements featured the
somewhat vague 'name' attribute, which was used inconsistently
throughout v0.1. This was combined with a 'type' attribute which
attempted to indicate whether an input or output was
"internal" to the module's language, or "external" as in a
file or URL. Output elements had 'ref' attributes to indicate
the file or URL address of a module's output, but input elements
had no ability to specify a particular resource using
'ref'. Module source elements also featured 'ref' attributes,
but this functioned as in indicator of a file to be read into
the module, and not an output produced by the module, as was the
case for the 'ref' attribute in output elements.
This project introduces a new element-type in OpenAPI v0.3,
which we have called 'vessel' elements. A vessel element is
designed to act as a pointer to a resource which a module either
consumes when executed, or produces as a consequence of its
execution. A vessel is required inside each module
input, output, or source
element. Instead of an input or output declaring a 'type', the
vessel element's name indicates the type of resource used;
internal-, file-, url-,
and script-type vessels have been implemented in
OpenAPI v0.3. The following code demonstrates how a module input
can refer to an internal resource through a vessel element:
<input name="in1">
<internal symbol="pricelist"/>
</input>
In OpenAPI v0.3 a module input or output element now has only
one attribute: 'name'. The 'name' attribute is used by OpenAPI
pipeline XML and an OpenAPI glue system to indicate a specific
input or output element; the 'name' attribute does not have to
correspond to the way a resource is labelled within a module
source script. Attributes of a module input or output's vessel
element will still correspond to details inside source scripts,
but a module author is no longer constrained by the naming
choices of a script author.
A vessel element serves as an interface to a resource which
is common across the parts of OpenAPI XML which consume or
produce resources. A vessel is not intended to be the resource
itself, but rather a pointer to the resource. In the case of
module inputs a vessel points to a resource which should already
exist at the time a module is executed; vessel elements in
module outputs point to a resource which will exist after a
module is executed; vessel elements in module
source elements point to a reference which will be
read in by an OpenAPI glue system in order to execute a
module.
A vessel type can be defined for any type of resource which
can be referenced by a pointer to the the resource, e.g. a file
path for file resources, or a URL for resources available over a
network interface. An OpenAPI glue system should provide methods
for resolving each type of vessel resource it supports; a glue
system does not have to be able to read from the resource
indicated by a vessel, but it must be able to determine whether
or not the resource indicated by a vessel can be accessed by a
module's source scripts.
Vessel use in module inputs and outputs
Under OpenAPI v0.1 module input and output elements could
have either an "internal" or "external" type; internal
inputs and outputs pass objects native to the module's language,
while external inputs and outputs pass external objects like
files or URLs. OpenAPI v0.3 replaces this mechanism with several
vessel elements for module inputs and outputs. The vessel types
available in OpenAPI v0.3 are internal,
file and url. A description of these
vessel types follows.
Internal vessels
For each language supported by an OpenAPI glue system there
is support for transmitting an object internal to this language
from one module to another through outputs and inputs with
internal vessel elements. An
internal-type vessel contains a 'symbol' attribute,
which indicates the symbol with which the object is associated
within a module's source scripts. The following XML demonstrates
how an internal-type input was described for a
module input in OpenAPI v0.1:
<module language="R">
<input name="prices" type="internal">
...
</input>
...
</module>
The following XML code demonstrates how this is now done
using an internal-type vessel in OpenAPI v0.3:
<module language="R">
<input name="prices">
<internal symbol="pricelist"/>
...
</input>
...
</module>
This XML describes a module to be executed using R, with an
input named 'prices'; an OpenAPI pipeline can describe a pipe
from another module to this input using this name. The 'prices'
input contains an internal-type vessel, which
indicates that the resource has the symbol 'pricelist'; this
indicates that the module's source scripts require an object to
be available with the symbol 'pricelist'. The input 'name' and
internal 'symbol' do not have to match, allowing the module
author to label elements within a pipeline however she chooses
without modifying the source scripts.
The next code example demonstrates how an internal-type
output is described in OpenAPI v0.1:
<module language="python">
...
<output name="suburbs" type="internal">
...
</output>
</module>
The following code demonstrates how an
internal-type vessel can be used with a module
output in OpenAPI v0.3:
<module language="python">
...
<output name="suburbs">
<internal symbol="suburbNames"/>
...
</output>
</module>
This XML describes a module to be executed using Python, with
an output named 'suburbs'; an OpenAPI pipeline can connect this
output to another module's input using this name. The 'suburbs'
input contains an internal-type vessel which
indicates that the resource has assigned to the symbol
'suburbNames'; this indicates that the module's source scripts
will have produced an object internal to the python session with
the symbol 'suburbName' during execution. As with module inputs,
an output 'name' and internal vessel 'symbol' can be completely
different, allowing a module author freedom in naming outputs
without altering source scripts.
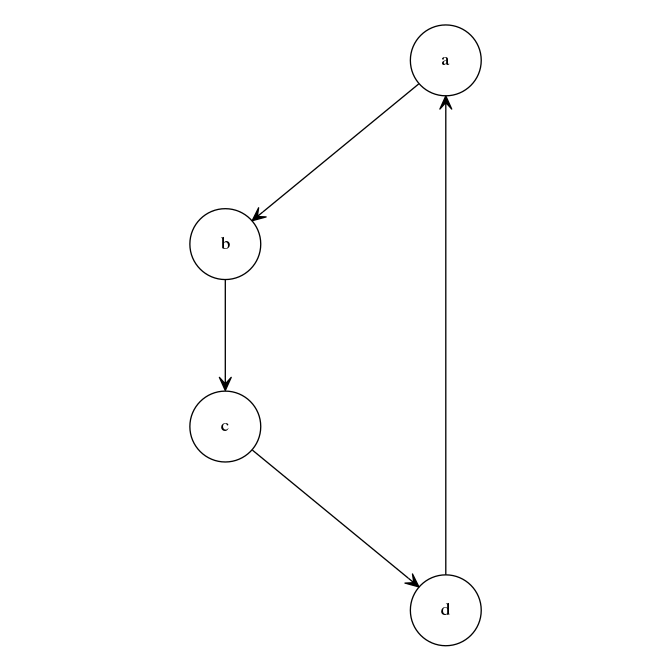
Example: names and symbols
The 'simpleGraph' pipeline
demonstrates a pipeline containing modules which use
internal vessel elements to pass objects between
modules. This example demonstrates how internal
vessels allow module input and output names to differ from the
symbols assigned to the object in the source scripts. The
'createGraph.xml' module file contains the following:
<?xml version="1.0"?>
<module xmlns="http://www.openapi.org/2014/" language="R">
<description>Create a simple directed graphNEL graph object with four nodes and four edges</description>
<source>
<script><![CDATA[library(graph)
nodes <- c("a", "b", "c", "d")
edgeList <- list(a=list(edges=c("b")),
b=list(edges=c("c")),
c=list(edges=c("d")),
d=list(edges=c("a")))
directedGraph <- new("graphNEL", nodes=nodes, edgeL=edgeList,
edgemode="directed")]]>
</script>
</source>
<output name="directedGraph">
<internal symbol="directedGraph"/>
<format formatType="text">R "graphNEL" object</format>
</output>
</module>
The source script of this module creates a "graphNEL"
object called 'directedGraph' in an R session. The module
declares an output, also called 'directedGraph', which uses an
internal type vessel to indicate that the object
with symbol name 'directedGraph' is available to other
modules.
The module file 'layoutGraph.xml' contains the following:
<?xml version="1.0"?>
<module xmlns="http://www.openapi.org/2014/" language="R">
<description>Lays out a graphNEL graph using the Rgraphviz package</description>
<input name="myGraph">
<internal symbol="myGraph"/>
<format formatType="text">R "graphNEL" object</format>
</input>
<source>
<script><![CDATA[library(Rgraphviz)
Ragraph <- agopen(myGraph, "myGraph")]]>
</script>
</source>
<output name="Ragraph">
<internal symbol="Ragraph"/>
<format formatType="text">R "Ragraph" object</format>
</output>
</module>
This module requires an input, named 'directedGraph' by the
module author. This input's internal vessel element
indicates that the source script requires this object to be
given the symbol 'myGraph' in the R environment where the source
script is executed. The pipeline file,
'simpleGraph-pipeline.xml', loads these two modules as
'createGraph' and 'layoutGraph' respectively, and connects the
output from the former to the input in the latter with the
following pipe:
<pipe>
<start component="createGraph" output="directedGraph"/>
<end component="layoutGraph" input="directedGraph"/>
</pipe>
File vessels
OpenAPI v0.1 described any module output or input object
which is not internal to the module language as "external"
type, which was the principal method for passing file resources
between modules. OpenAPI v0.3 allows for resources to be
transmitted between module outputs and inputs via the filesystem
using file-type vessel elements. A
file vessel element contains a 'ref' attribute and
an optional 'path' attribute; the 'ref' attribute contains a
file path which indicates where the file object is located; the
optional 'path' attribute indicates a file path from which a
glue system should search for a file matching the value in
'ref'.
Module inputs which require file resources are described in
OpenAPI v0.1 as follows:
<module language="R">
<input name="suburbIDs" type="external">
...
</input>
...
</module>
Module inputs which require file resources are described in
OpenAPI v0.3 as in the following:
<module language="R">
<input name="suburbIDs">
<file ref="151023-table.csv"/>
...
</input>
...
</module>
This XML describes a module with an input named 'suburbIDs'
which requires a file to be available at the file path
'151023-table.csv'. The input 'name' is used by the glue system
to identify this particular module input, and is not required to
match any objects or symbols in the source scripts. A module
author can use an input with a file vessel to
indicate that a module requires a file with a specific name, but
can refer to it descriptively within her pipeline without
altering the source script.
The previous example named a relative file path, indicating
that the module source scripts require a file resource be
available at the file path provided, relative to the directory
in which the module scripts are executed—a glue system is
responsible for ensuring this file resource is available in the
correct location when the module is executed; this is the
situation when an input requires a file resource which is itself
an output from another module. The new specification of
file vessels also allows module inputs to indicate
file resources with absolute file locations, as in the following
example:
<module language="R">
<input name="suburbIDs">
<file ref="/home/biguser/datasets/matches/151023-table.csv"/>
...
</input>
...
</module>
This example demonstrates a module which requires a file
resource from from the local filesystem, available at the file
path '/home/biguser/datasets/matches/151023-table.csv'. As the
file path provided is absolute the glue system does not need to
resolve the input resource's location—this module can
receive a file resource as an input without that resource coming
from the output of another module. This allows a module to be
executed in isolation, and allows for pipelines to be started from
data stored in files.
OpenAPI v0.1 described module outputs which reference file
resources as follows:
<module language="R">
...
<output name="birdPicture" type="external" ref="Rplots.pdf">
...
</output>
</module>
Using a file-type vessel OpenAPI v0.3 describes
a module output which produces a file resource as:
<module language="R">
...
<output name="birdPicture">
<file ref="Rplots.pdf"/>
...
</output>
</module>
This XML describes a module with an output named
'birdPicture'. This name is used by an OpenAPI glue system to
refer to the a file resource produced by this module, and to
provide this resource to other modules' input in a pipeline; the
file resource indicated by this output has the filename
'Rplots.pdf'. The name given to the module output does not have
to match the name of any object or resource in or produced by
the module's source scripts, allowing a module author to name
outputs as she likes without altering the source scripts. The
file vessel's 'ref' attribute names the file path
of a resource produced on execution of the module—the file
path given by 'ref' should match the file path of a file
resource produced by the source script. A glue system must be
able to resolve the location of an absolute file path given by
'ref'.
It is also possible for the file-type vessel of
a module output to provide an absolute file path in 'ref':
<module language="python">
...
<output name="vendors">
<file ref="/data/plots/vendors/processed.csv"/>
...
</output>
</module>
This XML example indicates that the module source scripts will
produce a file resource at the file path
'/data/plots/vendors/processed.csv'.
Example: passing external objects into modules
The introduction of a file vessel element makes
it much easier for a module author to pass file objects
(previously "external" type inputs) to module source
scripts. The 'shellExample'
pipeline provides an example of this. The module file
'plotgraphviz.xml' follows:
<?xml version="1.0"?>
<module xmlns="http://www.openapi.org/2014/" language="shell">
<description/>
<input name="dotfile">
<file ref="dot.dot"/>
<format formatType="text">dot file</format>
</input>
<source>
<script><![CDATA[dot -Tpng dot.dot -o simpleGraph.png]]></script>
</source>
<output name="simpleGraph.png">
<file ref="simpleGraph.png"/>
<format formatType="text">PNG file</format>
</output>
</module>
This module requires a file resource, 'dot.dot', to be
available in the working directory when the module source script
is executed. With the use of a file vessel element
we can see that the module author can name the input anything
she likes—'dotfile' in this case—and use the file
vessel element to indicate the name of the file required by the
script.
The introduction of file vessel elements also
means it is possible for a file resource provided as another
module's input to have an entirely different filename to the
resource required as a module input. Consider module file
'graphtodot.xml':
<?xml version="1.0"?>
<module xmlns="http://www.openapi.org/2014/" language="R">
<description/>
<input name="directedGraph">
<internal symbol="directedGraph"/>
<format formatType="text">R "graphNEL" object</format>
</input>
<source>
<script><![CDATA[library(Rgraphviz)
toDot(directedGraph, "directedGraph.dot")]]></script>
</source>
<output name="dotfile">
<file ref="directedGraph.dot"/>
<format formatType="text">dot file</format>
</output>
</module>
This module produces an output named 'dotfile' which
references a file resource with the filename
'directedGraph.dot'. The pipeline can use 'directedGraph.dot' as
the required 'dot.dot' by connecting the module output and input
via a pipe:
<pipe>
<start component="graphtodot" output="dotfile"/>
<end component="plotgraphviz" input="dotfile"/>
</pipe>
Example: starting with an external input
The introduction of file vessel elements also
allows a module or pipeline to be started from a file
resource. The 'fileStart'
module demonstrates how a module can require an input where
the file vessel element indicates an absolute file path:
<?xml version="1.0"?>
<module xmlns="http://www.openapi.org/2014/" language="R">
<description>Read the iris data from a CSV file and produce a pie chart of the species</description>
<input name="datafile">
<file ref="~/Desktop/iris.csv"/>
<format formatType="text">CSV file</format>
</input>
<source>
<script><![CDATA[data <- read.csv("~/Desktop/iris.csv")
speciesTable <- table(data$Species)
png()
pie(speciesTable)
dev.off()]]></script>
</source>
<output name="species">
<file ref="Rplot001.png"/>
<format formatType="text">PNG file</format>
</output>
</module>
The input, named 'datafile', references a file at path
'~/Desktop/iris.csv'. The module source script also uses this full
file path. In this way a module author can start a module, and a
whole pipeline, from a file resource on her local machine rather
than just from a file resource which results from a module
output.
URL vessels
Though undocumented, the OpenAPI v0.1 supported the
transmission of resources referenced by URLs by declaring a
module output or input with an "external" type. OpenAPI v0.3
introduces the capacity to transmit resources referenced by URLs
between modules using url-type vessel elements. A
url vessel element contains a 'ref' attribute,
which contains a URL at which a resource can be resolved over
the web via HTTP or HTTPS, and some other network protocols.
The use of a url type vessel for a module input
is demonstrated below:
<module language="R">
<input name="allEpisodes">
<url ref="https://en.wikipedia.org/wiki/List_of_The_X-Files_episodes"/>
...
</input>
...
</module>
This examples describes a module with an input named
'allEpisodes'. The module source scripts require that a resource
be available over HTTPS at the URL given in the url
vessel element's 'ref'
attribute—'https://en.wikipedia.org/wiki/List_of_The_X-Files_episodes'.
Such a module could be executed in isolation, or could serve as
the start of a pipeline as it requires as input an already
existing resource which can be used to start a pipeline.
A module which produces a resource available over the web at
a URL can describe this output using a url vessel
as follows:
<module language="R">
...
<output name="episodeTable">
<url ref="http://127.0.0.1:80/conduit/episodes.html"/>
...
</output>
</module>
This XML describes a module which will produce a resource
available over the web at the URL
'http://127.0.0.1:80/conduit/episodes.html'. This output is given
the name 'episodeTable'. The output name can be used by an OpenAPI
glue system to pass this output resource to other modules' inputs
through a pipeline.
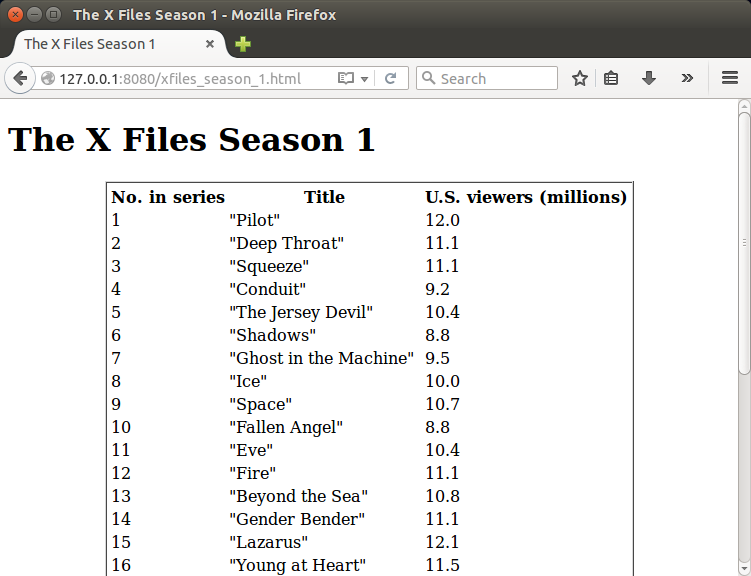
Example: URL vessels
The 'xfiles_pipeline'
pipeline demonstrates how a module can use a
url vessel element in a module input. The module
file 'xfiles_season1.xml':
<?xml version="1.0"?>
<module xmlns="http://www.openapi.org/2014/" language="R">
<description/>
<input name="wikiurl">
<url ref="https://en.wikipedia.org/wiki/List_of_The_X-Files_episodes" />
<format formatType="text">HTML file</format>
</input>
<source>
<script><![CDATA[library(RCurl)
library(XML)
url <- getURL("https://en.wikipedia.org/wiki/List_of_The_X-Files_episodes")
html <- htmlParse(url)
tables <- readHTMLTable(html)
season1 <- tables[[2]]
names(season1) <- gsub("
", " ", names(season1))
season1 <- season1[c(1, 3, 8)]
season1[,1] <- as.numeric(as.character(season1[,1]))
season1[,2] <- as.character(season1[,2])
season1[,3] <- gsub("[[][0-9].]$", "", season1[,3])
season1[,3] <- as.numeric(season1[,3])]]></script>
</source>
<output name="season1">
<internal symbol="season1"/>
<format formatType="text">R data frame</format>
</output>
</module>
This module has an input, named 'wikiurl', which requires a
URL resource resolvable at
'https://en.wikipedia.org/wiki/List_of_The_X-Files_episodes'.
The module can be started from this URL, as in the example of
starting a module from a file resource above.
This pipeline also contains a module which produces a URL
output, 'season1_html.xml':
<?xml version="1.0"?>
<module xmlns="http://www.openapi.org/2014/" language="R" host="conduit@127.0.0.1:2222">
<description/>
<input name="season1">
<internal symbol="season1"/>
<format formatType="text">R data frame</format>
</input>
<source>
<url ref="http://127.0.0.1:8080/urlTesting/season1_html.R"/>
</source>
<output name="season1_html">
<url ref="http://127.0.0.1:8080/xfiles_season_1.html"/>
<format formatType="text">html file</format>
</output>
</module>
This module produces an output, 'season1_html', which can be
accessed via the URL
'http://127.0.0.1:8080/xfiles_season_1.html'. This output could
also be passed to the input of another module requiring a URL
resource though a pipe in a pipeline.
Vessel use in module sources
OpenAPI v0.1 described two methods for providing source
script through module source elements: by inline scripts, and by
reference. OpenAPI v0.3 replaces both methods with the use of a
vessel element to indicate module source scripts. The
file and url vessel elements described
above can both be used inside module source
elements to indicate a resource. The 'season1_html.xml'
module file provides a source script using a url
vessel:
<?xml version="1.0"?>
<module xmlns="http://www.openapi.org/2014/" language="R" host="conduit@127.0.0.1:2222">
<description/>
<input name="season1">
<internal symbol="season1"/>
<format formatType="text">R data frame</format>
</input>
<source>
<url ref="http://127.0.0.1:8080/urlTesting/season1_html.R"/>
</source>
<output name="season1_html">
<url ref="http://127.0.0.1:8080/xfiles_season_1.html"/>
<format formatType="text">html file</format>
</output>
</module>
In this example the module which uses the R language wraps a
source script resource available at the URL
'http://127.0.0.1:8080/urlTesting/season1_html.R'.
In general a module source should be able to consume a
resource from any type of vessel which can reference a text
document. Module sources cannot contain internal
type vessels as there is no native internal object defined for
the OpenAPI architecture.
Inline source scripts are also described using a vessel
element, script, described below.
Script vessels
In OpenAPI v0.1 source scripts could be provided directly as
the content of module source elements, as in the
following example:
<module language="R">
...
<source><![CDATA[rent.lm <- lm(price ~ suburb + beds, data = aucklandListings)]]>
</source>
...
</module>
OpenAPI v0.3 introduces the script-type vessel
element with which to wrap source scripts inline within a
module's source element. The script element has no
attributes, and contains the text source script for use in the
module. If the source script contains line breaks or XML control
characters it can be wrapped in a CDATA block to preserve
these. The module file 'createGraph.xml' uses a script vessel
element to wrap an inline source script:
<?xml version="1.0"?>
<module xmlns="http://www.openapi.org/2014/" language="R">
<description>Create a simple directed graphNEL graph object with four nodes and four edges</description>
<source>
<script><![CDATA[library(graph)
nodes <- c("a", "b", "c", "d")
edgeList <- list(a=list(edges=c("b")),
b=list(edges=c("c")),
c=list(edges=c("d")),
d=list(edges=c("a")))
directedGraph <- new("graphNEL", nodes=nodes, edgeL=edgeList,
edgemode="directed")]]>
</script>
</source>
<output name="directedGraph">
<internal symbol="directedGraph"/>
<format formatType="text">R "graphNEL" object</format>
</output>
</module>
Vessel use in pipelines
Version 0.1 of OpenAPI allowed pipeline authors to reference
the contents of a pipeline component in one of two way: by
module or pipeline XML wrapped in component
element, or by using the component's ref attribute
to indicate a file containing the XML. OpenAPI v0.3 retains the
use of inline XML, but replaces the use of the ref
with the use of vessel elements. The 'test_pipeline' pipeline
uses file vessel to reference files containing
OpenAPI module XML:
<?xml version="1.0"?>
<pipeline xmlns="http://www.openapi.org/2014/">
<description/>
<component name="module1" type="module">
<file ref="module1.xml"/>
</component>
<component name="module2" type="module">
<file ref="module2.xml"/>
</component>
<pipe>
<start component="module1" output="x"/>
<end component="module2" input="numbers"/>
</pipe>
</pipeline>
When a vessel element is used to reference a pipeline or
module XML document the parent component must
provide a type attribute indicating that the XML
document is either a "module" or "pipeline". As of OpenAPI
v0.3 either a file or url vessel can
be used to indicate an XML document for use as a pipeline
component.
It is still possible to provide pipeline or module XML
inline. No type attribute is required when inline
XML used, as in the example below:
<pipeline>
<component name="m1">
<module language="R">
<source><script>x <- rnorm(10)</script></source>
<output name="x">
<internal symbol="x"/>
<format formatType="text">R numeric vector</format>
</output>
</module>
</component>
</pipeline>
Module language
Module elements in OpenAPI v0.1 contained
platform elements which described the platform in
which the module's source scripts were to be executed. A module
which wrapped source scripts in the R language was described as
follows:
<module>
<platform name="R/>
...
</module>
OpenAPI v0.3 removes the platform element, and
replaces it with a 'language' attribute in module
elements. The use of "language" better makes explicit what is
intended by a module wrapping source scripts: all of a module's
source scripts are written in a programming language, e.g. R,
Python, or shell. An OpenAPI glue system must be able to execute
the code contained in these source scripts, but it is up to the
glue system author how this execution is carried out. Thus,
while the language of a module's source scripts should be fixed,
the platform in which the scripts are executed can vary. OpenAPI
v0.3 describes a module's language as follows:
<module language="python">
...
</module>
A module element must declare a language. Hinton
and Murrell said of OpenAPI v0.1 that a glue system "must be
able to execute the code specified by a module's
source elements in the platform specified by the
module" (2015a). For
OpenAPI v0.3, a glue system must be able to execute the code
specified by a module's source elements in the
'language' specified by the module; the platform of
execution is up the to the glue system. A glue system should
still report if it does not support a module's stated
'language'.
Module host
One of the limitations identified in OpenAPI v0.1 was its
inability to support various system and software version
dependencies of modules. OpenAPI v0.3 implements support for
module hosts—machines on which a module is guaranteed to
execute properly—to address this issue.
A module host is a machine which meets the system, software,
and version requirements of a module's sources scripts. A host
machine should be available to an OpenAPI glue system over a
network. The initial implementation of module hosts described in
this project expects that the host machine will provide a Linux-
or UNIX-like shell environment over the Secure Shell (SSH)
network protocol.
An OpenAPI glue system is responsible for connecting with a
module's host machine. The glue system should support passing
module outputs to a module being executed on a host machine, and
executing the module's source scripts on the remote host. A glue
system is also responsible for resolving output object locations
produced on the remote host and providing these as inputs to other
modules, either locally or on other host machines.
A module's host is declared through the optional 'host'
attribute in a module element. The 'host' attribute
should contain a Uniform Resource Identifier (URI) which
identifies a networked machine on which the module can be
correctly executed. The implementation of module hosts described
in this project assumes the SSH scheme. The URI for the host
should have the form username@host:port. For
example:
<module language="R" host="conduit@openapi.org:2222">
...
</module>
This XML describes a module whose source scripts will be
executed on the host a host machine at the address 'openapi.org',
accessed via port '2222', logged in as the user 'conduit'.
The SSH protocol requires that a user's connection be
authenticated through some method. The improvements to conduit
version 0.3 support authentication using an SSH identity
keyfile. Authentication via password could also be supported by
including a password in the host URI, but this has not been
implemented in the improvements to conduit v0.3.
As a module's host machine is accessed using the SSH protocol
the machine can be provided in any manner which provides an SSH
interface. A glue system does not have to be responsible for
provisioning a module's environment, only for access to a host
via SSH. A host machine can of course be a physical machine, but
it can also be provided by a virtual machine
environment. Possible candidates for providing host machines
include Docker Engine and Vagrant. Docker Engine is a software
application for creating and running Docker
containers—wrappers for software applications which can be
run in Linux and accessed via SSH (Docker, 2015). Vagrant is an
application for creating, configuring and running virtual
machine environments, which can also be accessed via SSH (Vagrant, 2014). Configuration
files for host machines provided via virtual machines or
containers can be easily shared. This means that a module author
could share a configuration file, allowing OpenAPI users to
provision a module's host machine on their own machine, instead
of a module author having to provide access to a machine to
module users in perpetuity. The technical requirements
appendix describes how to provision a Vagrant machine to be
used as a module host machine for many of the example modules in
the pipeline examples
appendix.
Example: executing a module on a host machine
The 'kmeans.xml' module file is part of the 'irisplots_host'
pipeline. The module XML code follows:
<?xml version="1.0"?>
<module xmlns="http://www.openapi.org/2014/" language="R" host="conduit@127.0.0.1:2222">
<description/>
<input name="data">
<internal symbol="data"/>
<format formatType="text">R data frame</format>
</input>
<source>
<script><![CDATA[clusters <- kmeans(data, 3)]]></script>
</source>
<output name="clusters">
<internal symbol="clusters"/>
<format formatType="text">R kmeans object</format>
</output>
</module>
This module makes use of a host machine,
'conduit@127.0.0.1:2222'. When a glue system executes this module
it will do so on the host machine at the address provided,
accessed via SSH. The glue system will ensure that the input,
'data', is available to the module source script on the host
machine. The glue system will also make available the output,
'clusters', to any modules on the local machine where the glue
system is called, or any subsequent host machines called by
modules.
Example: meeting module dependencies
The 'xfiles_season1.xml' module file in the 'xfiles_pipeline' uses a
module host machine to meet the dependencies of the host
script. The module XML follows:
<?xml version="1.0"?>
<module xmlns="http://www.openapi.org/2014/" language="R" host="conduit@127.0.0.1:2222">
<description/>
<input name="season1">
<internal symbol="season1"/>
<format formatType="text">R data frame</format>
</input>
<source>
<url ref="http://127.0.0.1:8080/urlTesting/season1_html.R"/>
</source>
<output name="season1_html">
<url ref="http://127.0.0.1:8080/xfiles_season_1.html"/>
<format formatType="text">html file</format>
</output>
</module>
This module produces a resource which is available over HTTP at
the URL 'http://127.0.0.1:8080/xfiles_season_1.html'. Making a
resource available over HTTP requires a web server—the host
machine given by 'conduit@127.0.0.1:2222' provides a web server
which can be accessed by the source script when executed on this
host. By using a host machine to execute this module's source
script the module user is saved the potentially complicated setup
of a web server.
The source script for this module—also hosted on the
web server of the host machine—contains the following R
code:
library(R2HTML)
outfile <- HTMLInitFile(outdir = "/var/www/conduit",
filename = "xfiles_season_1",
Title = "The X Files Season 1")
HTML("<h1>The X Files Season 1</h1>", outfile)
HTML(season1, outfile, row.names = FALSE)
HTML("Source: <a href=\"https://en.wikipedia.org/wiki/List_of_The_X-Files_episodes\">https://en.wikipedia.org/wiki/List_of_The_X-Files_episodes</a>",
outfile)
HTMLEndFile(outfile)
From the source script we can also see that the module requires
the R2HTML package to be installed in R. The host
machine has the R2HTML package available, and has
permitted the 'conduit' user to make changes to the web server's
file directory, '/var/www/conduit'.
XML validation
OpenAPI v0.3 has been described in XML Schema files for use
in validating pipeline and module XML. These XML Schema
documents have been implemented in the version 0.3 of the
conduit package (Hinton,
2016), as described in this report, to check the validity
of module and pipeline XML files loaded into Conduit.
The XML Schema files for OpenAPI version 0.3 modules and
pipelines can be found on the OpenAPI Validation repository at
https://github.com/anhinton/openapiValidation/releases/tag/v0.3.
Running 'wild' source scripts
One of the OpenAPI architecture's aims is to provide a simple
way of wrapping, connecting and executing existing data
scripts—'wild' scripts—without script authors having
written their code with OpenAPI in mind. This is similar to
Berners-Lee's original proposal for what would become the world
wide web, in which he insisted that hypertext would only succeed
if it could access existing data. Berners-Lee wrote of
hypertext, "what is required is a gateway program which will
map an existing structure onto the hypertext model" (1989). OpenAPI, too,
intends to map existing data sets, data analysis code,
workflows, and outputs onto the OpenAPI architecture. However,
as discussed in an earlier section, version 0.1 of the OpenAPI
architecture had several problems which made it difficult to
easily wrap 'wild' source scripts. This section demonstrates how
the changes in OpenAPI version 0.3 make it possible to meet this
aim.
Wrapping and subsetting a simple source script
In an earlier section we described an R script taken from the
STATS 330 course at the University of Auckland (Klaere, 2015). This script file
was distributed through the course website for students to
examine and execute the code used by Klaere in his lecture
slides. The first 32 lines of this script follow:
##############################
### STATS 330/762 - Lecture 3: Graphics
### 23/07/2015
### Dr. Steffen Klaere
### Supplementary Code
### NEEDS: hb1.csv and fever_data.csv
##############################
### Change working directory
# Adapt to your own needs
setwd("<Your Directory>")
### Packages
library(R330)
###################################
### First example, Exchange rates
###################################
exchange <- read.csv("hb1.csv")
exchange[,1] <- as.Date(sapply(exchange[,1],function(x){paste(1,x)}),format="%d %B %Y")
plot(USD~month,data=exchange,type="l",lwd=4,col="steelblue",xlab="Date",ylab="Exchange rate",main="Exchange rate between US$ and NZ$")
### Monthly rate of change
diff.in.logs <- sapply(2:nrow(exchange),function(i){log(exchange[i,2])-log(exchange[i-1,2])})
xvec <- seq(-0.2,0.1,length=100)
# Draw histogram
# density plots
pdf("data_dens.pdf",width=8,height=6)
hist(diff.in.logs,nclass=20,freq=F)
lines(density(diff.in.logs),col="blue",lwd=2)
dev.off()
The following module XML code shows how this script can be
wrapped in an OpenAPI v0.3 module file:
<?xml version="1.0"?>
<module xmlns="http://www.openapi.org/2014/" language="R">
<description>Run the first example from STATS 330 lecture 3, returning the first two plots as PNG files</description>
<input name="source_script">
<file ref="~/Desktop/330_Lecture3_2015.R"/>
<format formatType="text">R script</format>
</input>
<input name="data">
<file ref="~/Desktop/hb1.csv"/>
<format formatType="text">CSV file</format>
</input>
<source>
<file ref="subsettingScript.R"/>
</source>
<output name="exchange_rate">
<file ref="exchange_rate.png"/>
<format formatType="text">PNG file</format>
</output>
<output name="data_dens">
<file ref="data_dens.png"/>
<format formatType="text">PNG file</format>
</output>
</module>
The original script file contains a data input step, at line
19, where the CSV file 'hb1.csv' is read into the R session. The
module XML above references this CSV file as one of its inputs
using a file-type vessel element. This input
has been given the name 'data', and its file-type
vessel indicates it will consume a file resource found on the
local machine at '~/Desktop/hb1.csv'.
The original script produces—when executed in batch
mode—PDF files of plots at line 21 and at lines
29–32. However, the author of this module XML would like
to produce PNG files rather than PDF files, which means the
source file must be changed. Rather than edit the source script
directly, however, the use of module input vessel elements
allows the module author to operate on the source script using
the language in which the script is written. In the XML code
above the module author names a module input,
'source_script', which uses a file-type vessel
element to indicate that the module requires a file resource at
'~/Desktop/330_Lecture3_2015.R'. The module XML
above also contains a source element which requires a file
resource at 'subsettingScript.R'. The contents of
'subsettingScript.R' follow:
full_script <- readLines("~/Desktop/330_Lecture3_2015.R")
full_script[19] <- "exchange = read.csv(\"~/Desktop/hb1.csv\")"
full_script[21] <- paste("png(\"exchange_rate.png\")",
full_script[21],
"dev.off()",
sep = "; ")
full_script[29] <- "png(\"data_dens.png\")"
subset <- full_script[16:33]
eval(parse(text = subset))
The orignal R script, '330_Lecture3_2015.R', referenced
by the module input 'source_script', is consumed in line 1 of
the 'subsettingScript.R' script, and given the symbol
'full_script'. The 'full_script' object is then modified
in the following ways: the data step line in the original source
is modified to consume the local file referenced by the module
input 'data'; the plotting step at line 21 of the
'source_script' is wrapped in new code to produce a PNG file
when executed; and the code which produces a PDF file on line 29
of 'source_script' is modified to produce a PNG file. Lines
16–33 of the altered 'source_script' are given the
symbol 'subset'. Finally this subset of the original
'source_script' is executed using the eval
function.
When executed, the module source script given in
'subsettingScript.R' will produced two PNG
files—'exchange_rate.png' and 'data_dens.png'. The
module XML above indicates that the module will produce these
files by naming two outputs, 'exchange_rate' and
'data_dens', each of which references the filenames of the
PNG files produced using file-type vessel
elements.
This example demonstrates how the file-type
vessel elements introduced in OpenAPI version 0.3 make it easier
to wrap a 'wild' source script in an OpenAPI module. A
file-type vessel element is also used to indicate
the 'wild' script as one of the module's inputs—doing so
allows a module author to make changes to the source script
without having to create an altered copy of the script. This
method allows a module author to modify, subset, and customise
original source scripts to suit the module being written.
The full module and pipeline XML for this example, can be
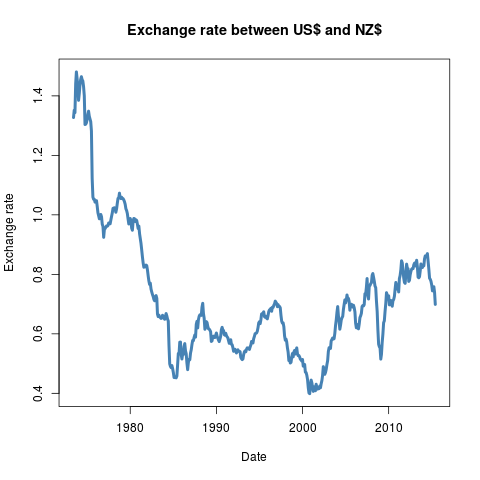
found in the 'exchangeRates'
section. The pipeline produces the graphic seen in figure 2. The 'exchangeRates' pipeline was
executed in conduit v0.3 with the following code:
library(conduit)
exchangeRates <- loadModule(
name = "exchangeRates",
ref = "examples/subsetting/exchange_rates.xml")
outputs <- runModule(exchangeRates)
The next section examines 'wild' scripts which are not made
available as simple script files.
Wrapping a blog post
One of the major difficulties with wrapping 'wild' scripts in
OpenAPI is that often data analysis code is not shared as a
script file—rather it is embedded and discussed on web
pages and in blog posts. The following example demonstrates how
the code in a blog post at
http://spatial.ly/2015/03/mapping-flows/—see figure 3 for a screen capture of the blog
post—can be captured and exectuted by OpenAPI v0.3.
The pipeline example below wraps and executes the code in a
blog post called 'Mapping Flows in R', which was posted at
http://spatial.ly/2015/03/mapping-flows/ (Cheshire, 2015). At the time
writing the pipeline there was a technical issue with the site
hosting the post which meant the R code was displayed
incorrectly, so a blog network feed syndicating the post was
used in the pipeline
instead—http://blogs.casa.ucl.ac.uk/category/r-spatial/.
As if to demonstrate the ephemeral nature of the web the
syndicating page changed its format at the time of this report
being written, which led to the example below to be based on an
archived copy of the blog post. The original code for this
pipeline can be found in the 'ukCommuter' section; the code
used in this example can be found in the 'ukCommuterErsatz'
section.
The following XML file 'extract_scripts.xml' shows how the blog
post is wrapped in a module:
<?xml version="1.0"?>
<module xmlns="http://www.openapi.org/2014/" language="R">
<description>extract R code from blog post http://spatial.ly/2015/03/mapping-flows/ (actually from mirror at http://127.0.0.1:8080/report/R%20spatial%20|%20The%20CASA%20Blog%20Network.html)</description>
<input name="blog">
<url ref="http://127.0.0.1:8080/report/R%20spatial%20|%20The%20CASA%20Blog%20Network.html"/>
<format formatType="text">html file</format>
</input>
<source>
<script><![CDATA[library(XML)
library(RCurl)
## extract source article from category feed
source_lines <-
readLines("http://127.0.0.1:8080/report/R%20spatial%20|%20The%20CASA%20Blog%20Network.html")
heading <- grep("<h1 class=\"entry-title\">[[:print:]]*http://spatial.ly/2015/03/mapping-flows/", source_lines)
start <- grep("^[ ]*<article", source_lines)
start <- start[start < heading]
start <- start[length(start)]
end <- grep("</article>", source_lines)
end <- end[end > start][1]
source_lines <- source_lines[start:end]
## save a local copy of the article
writeLines(source_lines, "source_lines.txt")
## extract code chunks
code_start <- grep("<code>", source_lines)
code_end <- grep("</code>", source_lines)
code_lines <- lapply(seq_along(code_start),
function (i, start, end) {
seq(start[i] + 1, end[i] - 1)
}, code_start, code_end)
code <- lapply(code_lines,
function(x, src) {
c(gsub("<br />", "", src[x]), "")
}, source_lines)
## remove rogue tags and replace characters
code <- lapply(X=code, FUN=gsub, pattern="<", replace="<")
code <- lapply(X=code, FUN=gsub, pattern=">", replace=">")
code <- lapply(X=code, FUN=gsub, pattern="<br>", replace="")
## save code chunks as R scripts
data_script <- unlist(code[2:5])
writeLines(data_script, "data_script.R")
graphics_script <- unlist(code[c(1, 6:10)])[-c(1, 3)]
graphics_script <-
c(graphics_script, "ggsave(file = \"ukCommuterGraphic.png\")")
writeLines(graphics_script, "graphics_script.R")]]></script>
</source>
<output name="wu03ew_v1.csv.zip">
<url ref="http://127.0.0.1:8080/report/wu03ew_v1.csv.zip"/>
<format formatType="text">zip file</format>
</output>
<output name="msoa_popweightedcentroids.csv">
<url ref="http://127.0.0.1:8080/report/msoa_popweightedcentroids.csv"/>
<format formatType="text">CSV file</format>
</output>
<output name="raw_source">
<file ref="source_lines.txt"/>
<format formatType="text">txt file</format>
</output>
<output name="data_script">
<file ref="data_script.R"/>
<format formatType="text">R script</format>
</output>
<output name="graphics_script">
<file ref="graphics_script.R"/>
<format formatType="text">R script</format>
</output>
</module>
This module has one input, named 'blog', which requires a
resource available at the URL
'http://127.0.0.1:8080/report/R%20spatial%20|%20The%20CASA%20Blog%20Network.html'.
The module source consumes the file at this URL, and writes a
copy of this file to disk as 'source_lines.txt'. The module
source script extracts the lines of code from the blog post and
tidies the code for execution. The code is split into two
objects, called 'data_script' and 'graphics_script'. These
objects are saved as files 'data_script.R' and
'graphics_script.R' respectively. The three files created by
the module source are named as module outputs 'raw_source',
'data_script', and 'graphics_script' respectively. The
module also names two other outputs, 'wu03ew_v1.csv.zip' and
'msoa_popweightedcentroids.csv', which reference the URLs of
two data files required by the code in the blog post.
By using a module input with a url-type vessel
element indicating the original blog post OpenAPI 0.3 allows a
module author to not only wrap and execute the code contained in
a blog post, but also to maintain a connection to the original
blog post. In this way a module can be updated when its input
file is updated. Producing the source code of the original blog
resource as an output allows a module author to preserve a blog
post as it was when the code was executed. Passing the extracted
code out as module outputs allows the module author to preserve
the code content of a blog post in an executable form, which can
now be passed to another module.
The original blog post mentions that the resource called
'wu03ew_v1.csv.zip' in the module XML above needs to be
unzipped for the code to be executed. The module XML file
'extract_flow_data.xml' extracts this data from its zip
file:
<?xml version="1.0"?>
<module xmlns="http://www.openapi.org/2014/" language="R">
<description>Download UK flow data and unzip to make available as local file - http://127.0.0.1:8080/report/R%20spatial%20|%20The%20CASA%20Blog%20Network.html</description>
<input name="wu03ew_v1.csv.zip">
<url ref="http://127.0.0.1:8080/report/wu03ew_v1.csv.zip"/>
<format formatType="text">zip file</format>
</input>
<source>
<script><![CDATA[download.file("http://127.0.0.1:8080/report/wu03ew_v1.csv.zip", "wu03ew_v1.csv.zip")
unzip("wu03ew_v1.csv.zip", files = "wu03ew_v1.csv")]]></script>
</source>
<output name="data">
<file ref="wu03ew_v1.csv"/>
<format formatType="text">CSV file</format>
</output>
</module>
The 'ukCommuterErsatz' pipeline connects the
'wu03ew_v1.csv.zip' output from the module file
'extract_scripts.xml' to the corresponding input of the above
module, which then returns the uncompressed CSV file as a module
output, named 'wu03ew_v1.csv'. The data scripts embedded in
the blog post should now have all the inputs required for
execution. A module XML file, 'set_coords.xml', receives and
executes the 'data_script' output from
'extract_scripts.xml' above, and the
'uk_flow_graphic.xml' module XML file executes the
'graphics_script' output, producing the graphic shown in figure 4. The full details of this pipeline
can be seen in the 'ukCommuterErsatz'
section. The 'ukCommuterErsatz' pipeline is executed in
conduit v0.3 using the following code:
library(conduit)
ukCommuterErsatz <- loadPipeline("ukCommuterErsatz",
"examples/ukCommuterErsatz/pipeline.xml")
outputs <- runPipeline(ukCommuterErsatz, targetDirectory = tempdir())
The introduction of url- and
file-type vessel elements in OpenAPI v0.3 has made
it possible to "execute" a blog post, as in the example
above. Ordinarily the code in a blog post requires some
manipulation to execute, and it is almost never as simple as
copying, pasting, and running the code. As in the example here,
data analysis code in blog posts can be presented in fragments,
with data and software requirements described both pragmatically
in the code and in the text of the blog post itself, as with the
input files in this example. By wrapping a blog post in an
OpenAPI module the author provides an interface to a blog post
which allows the repeated execution of the code on different
machines. The module file 'extract_scripts.xml' in the
'ukCommuterErsatz' pipeline above provides access to not just
the blog post's code, but to its input files. This allows for
re-use, not just of the script, but of the inputs
themselves.
The 'ukResponseErsatz'
pipeline re-uses the input resources extracted from the blog
post in the 'ukCommuterErsatz' pipeline. The 'ukResponseErsatz'
pipeline wraps a blog post, 'Mapping Flows in R ... with
data.table and lattice' (Perpiñán Lamigueiro,
2015), which demonstrates another method of producing a
graphic like the one in Cheshire (2015) using the same data
sources—a screen grab of this blog post can be seen in figure 5. The 'ukResponseErsatz' pipeline
is derived from the 'ukResponse'
pipeline to preserve this example against the shifting sands
of online resources.
The module XML file 'extract_script.xml' follows:
<?xml version="1.0"?>
<module xmlns="http://www.openapi.org/2014/" language="R">
<description>extract R code from gist linked in blog post http://127.0.0.1:8080/report/Mapping%20Flows%20in%20R%20%E2%80%A6%20with%20data.table%20and%20lattice%20|%20Omnia%20sunt%20Communia!.html</description>
<input name="blog">
<url ref="http://127.0.0.1:8080/report/Mapping%20Flows%20in%20R%20%E2%80%A6%20with%20data.table%20and%20lattice%20|%20Omnia%20sunt%20Communia!.html"/>
<format formatType="text">html file</format>
</input>
<source>
<script><![CDATA[## extract gist url from blog
library(RCurl)
page <- readLines(textConnection(getURL("http://127.0.0.1:8080/report/Mapping%20Flows%20in%20R%20%E2%80%A6%20with%20data.table%20and%20lattice%20|%20Omnia%20sunt%20Communia!.html")))
line <- page[grep("<a href=\"http://127.0.0.1:8080", page)[1]]
gist_url <- strsplit(line, "href=\"")[[1]][2]
gist_url <- strsplit(gist_url, "/raw")[[1]][1]
## clone gist
local_path <- file.path(getwd(), "gist")
dir.create(local_path, recursive=TRUE)
system2("git", c("clone", gist_url, local_path))]]></script>
</source>
<output name="script">
<file ref="gist/mappingFlows.R"/>
<format formatType="text">R script</format>
</output>
</module>
This module has a single input, named 'blog', which indicates
a resource at the URL
'http://127.0.0.1:8080/report/Mapping%20Flows%20in%20R%20%E2%80%A6%20with%20data.table%20and%20lattice%20|%20Omnia%20sunt%20Communia!.html'
—the blog post. The source script of the module retrieves
the blog post, and locates a line in the post which refers to a
git repository where the post's code is available. The module
source script clones this repository, and labels a script of R
code as its sole output, named 'script'. As in the previous
example we have wrapped a blog post, this time one which shares
data analysis code by embedding the contents of a git
repository, and provided this script as an output for execution
in other modules. The 'ukResponseErsatz' pipeline passes the
inputs from the previous example and this data script to the
module 'mapping_flows.xml' for execution. The graphic
produced by this pipeline can be seen in figure 6. The code used to execute the
'ukRsponseErsatz' pipeline in conduit v0.3 follows:
library(conduit)
ukResponseErsatz <- loadPipeline("ukResponseErsatz",
"examples/ukResponseErsatz/pipeline.xml")
outputs <- runPipeline(ukResponseErsatz, targetDirectory = tempdir())
See 'ukResponseErsatz' for full
details of the module and pipeline XML used in this
example..
We have demonstrated how OpenAPI v0.3 allows module authors
to write modules which provide an interface to the resources
described in a blog post, and to re-use these resources as
inputs to another script, also provided by an interface to a
related blog post. Wrapping and labelling code and data
resources in OpenAPI modules and pipelines allows an author to
update her workflow as these online resources are updated. Using
OpenAPI as a wrapper around online resources makes them
executable in a way which is often not possible directly from a
web browser. OpenAPI modules and pipelines also present a method
for embedding and sharing data sets and data analyses online, as
it provides a means for readers to run and experiment with code,
and to extend data workflows.
Summary
The OpenAPI project offers a software solution to the problem
of helping people to connect with data. Version 0.1 of OpenAPI
had several problems in its specification which made it
difficult to author new modules and pipelines, and to wrap
existing data analysis scripts in OpenAPI modules. Vessel
elements, including file-, url-,
script-, and internal-type vessels,
were introduced in version 0.3 of OpenAPI. These elements
provide a uniform mechanism for referencing module input and
output resources, module source scripts, and pipeline
components. The use of vessel elements has made authoring
OpenAPI modules easier, and made OpenAPI modules better suited
to adapting 'wild' data analysis scripts. Version 0.3 of OpenAPI
also includes the introduction of module 'host' machines. These
machines provide a stable environment for executing module
source scripts which meet the module's software and version
dependencies. XML Schema for OpenAPI module and pipeline XML
files were also introduced in OpenAPI v0.3. These provide a
mechanism for ensuring that module and pipeline XML is
valid. The changes in OpenAPI v0.3 were implemented in version
0.3 of the R package conduit, a prototype OpenAPI glue
system.
The changes introduced in OpenAPI v0.3 have made it much
easier for module authors to wrap 'wild' data analysis scripts
which have not been written with OpenAPI in mind. This report
demonstrated several examples of how such existing scripts can
be wrapped and executed in OpenAPI. Several examples also
demonstrate how data analysis scripts can be subsetted and
modified for execution in OpenAPI in a way which preserves the
source material, and leverages the features of the script's
coding language.
Future work on the OpenAPI architecture could include
publishing a recommendation or specification for OpenAPI glue
systems, to assist glue system developers in implementing
OpenAPI features. More work also remains to be done in the
implementation of module 'host' machines, including implementing
a standard way for glue systems to authenticate connections with
host machines. The method for remote host authentication
implemented in OpenAPI and conduit versions 0.3 is not well
documented, and is not intended as a canonical model for all
host connections. It would be desireable to implement standard
interfaces for SSH keys and passwords. In particular, work
remains to be done on conduit's implementation of module
'host' machines, including the way input and output objects are
made available to various machines. There are also several other
potential candidates for vessel elements which could be
implemented in future versions of OpenAPI, including sockets and
websockets.
The conduit package is very much a work in progress, and
much remains to be implemented, documented, tested and improved
in future versions.
This project was funded in full by an Ockham Foundation
Postgraduate Scholarship in Statistics. The author expresses
his sincere thanks to the Ockham Foundation for making this
work possible.
References
Carey, V., Long, L., & Gentleman, R. (n.d.). RBGL: An interface to the BOOST graph library (Version 1.44.0). Retrieved from http://www.bioconductor.org
Gansner, E. R., & North, S. C. (2000). An open graph visualization system and its applications to software engineering. SOFTWARE - PRACTICE AND EXPERIENCE, 30(11), 1203–1233. Retrieved from http://www.graphviz.org/
Gentleman, R., Whalen, E., Huber, W., & Falcon, S. (n.d.). Graph: A package to handle graph data structures (Version 1.46.0).
Hansen, K. D., Gentry, J., Long, L., Gentleman, R., Falcon, S., Hahne, F., & Sarkar, D. (n.d.). Rgraphviz: Provides plotting capabilities for R graph objects (Version 2.12.0).
R Core Team. (2015). R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. Retrieved from https://www.R-project.org/
Technical requirements
The sections of R code demonstrated in this report were
executed in R version 3.2.3 (R Core Team,
2015) on an Ubuntu Linux 15.10 (Ubuntu, 2015) 64-bit machine.
Version 0.3 of the conduit package (Hinton, 2016) was used to
execute the pipeline examples in this report. The conduit
package requires the following R packages:
The machine used to execute the pipeline examples had
graphviz version 2.38.0 (Gansner
& North, 2000) installed. The following R packages
are also required to execute the example pipelines:
-
classInt (Bivand, 2015)
-
data.table (Dowle, Srinivasan, Short, &
Lianoglou, with contributions from Saporta & Antonyan,
2015)
-
git2r (Csardi,
Jefferis, Hester, Ram & Widgren, 2015)
-
ggplot2 (Wickham, 2009)
-
gridGraphviz (Murrell & Hinton,
2015)
-
lattice (Sarkar, 2008)
-
Rgraphviz (Hansen, Gentry, Long, Gentleman,
Falcon, Hahne & Sarkar, n.d.)
Several of the pipeline and modules example require a host
machine; this host machine is provide by a virtual machine
running Vagrant version 1.7.4 (Hashicorp, 2015) and
VirtualBox version 5.0.14 (Oracle Corporation, 2016). The
configuration files for this machine are available from
https://github.com/anhinton/vagrant-conduit/releases/tag/2015honsReport.
This machine can be installed and started using the following
commands in a Linux terminal:
## download machine configuration files
git clone https://github.com/anhinton/vagrant-conduit.git --branch 2015honsReport
## start host machine
cd vagrant-conduit
vagrant up
This may take some time on the first run, as Vagrant will
download a system image, and install the necessary packages on
the host machine. Once the vagrant machine has finished booting
for the first time you will need to add the machine's SSH key to
your ~/.ssh/known_hosts file for the pipeline examples
to work. This only needs to be done once, with:
## add host machine to known hosts
ssh-keyscan -H -p 2222 127.0.0.1 >> ~/.ssh/known_hosts
The host machine can be stopped with the command
vagrant halt, and started again with the command
vagrant up. More information on running Vagrant
machines can be found on the Vagrant website at
https://www.vagrantup.com/.
Pipeline examples
simpleGraph
Modules:
Pipeline: examples/simpleGraph/pipeline.xml
Execute pipeline in conduit
## Loading required package: methods
##
## Attaching package: 'conduit'
##
## The following object is masked from 'package:base':
##
## pipe
simpleGraph <-
loadPipeline(name = "simpleGraph",
ref = "examples/simpleGraph/pipeline.xml")
outputs <- runPipeline(simpleGraph)
file.exists("pipelines/simpleGraph/plotGraph/example.png")
shellExample
Modules:
Pipeline: examples/shellExample/pipeline.xml
Execute pipeline in conduit
library(conduit)
shellExample <-
loadPipeline(name = "shellExample",
ref = "examples/shellExample/pipeline.xml")
output <- runPipeline(shellExample)
file.exists("pipelines/shellExample/plotgraphviz/simpleGraph.png")
fileStart
Module: examples/fileStart/fileStart.xml
Execute module in conduit
library(conduit)
fileStart <-
loadModule(name = "fileStart",
ref = "examples/fileStart/fileStart.xml")
outputs <- runModule(fileStart, targetDirectory = "modules")
file.exists(outputs$species$object)
xfiles_pipeline
Modules:
Source script found at
http://127.0.0.1:8080/urlTesting/season1_html.R:
examples/xfiles_pipeline/season1_html.R
Pipeline:
examples/xfiles_pipeline/pipeline.xml
Execute pipeline in conduit
library(conduit)
xfiles_pipeline <-
loadPipeline(name = "xfiles_pipeline",
ref = "examples/xfiles_pipeline/pipeline.xml")
output <- runPipeline(xfiles_pipeline)
RCurl::url.exists(output$season1_html$season1_html$object)
irisplots_host
Modules:
Pipeline: examples/irisplots_host/pipeline.xml
Execute pipeline in conduit
library(conduit)
irisplots_host <-
loadPipeline(name = "irisplots_host",
ref = "examples/irisplots_host/pipeline.xml")
outputs <- runPipeline(irisplots_host)
file.exists(outputs$scattermatrix$scattermatrix.png$object)
ukCommuterErsatz
Modules:
Pipeline: examples/ukCommuterErsatz/pipeline.xml
Execute pipeline in conduit
library(conduit)
ukCommuterErsatz <- loadPipeline("ukCommuterErsatz",
"examples/ukCommuterErsatz/pipeline.xml")
outputs <- runPipeline(ukCommuterErsatz, targetDirectory = tempdir())
file.exists(outputs$uk_flow_graphic$pngfile$object)
ukResponseErsatz
Modules:
Pipeline: examples/ukResponseErsatz/pipeline.xml
Execute pipeline in conduit
library(conduit)
ukResponseErsatz <- loadPipeline("ukResponseErsatz",
"examples/ukResponseErsatz/pipeline.xml")
outputs <- runPipeline(ukResponseErsatz, targetDirectory = tempdir())
file.exists(outputs$mapping_flows$plot$object)